Weekly Advisor Meeting
Today’s Meeting is all about mussel reproduction, environmentally induced sex determinants and whatever is up with maternal and paternal mtDNA… cool stuff.
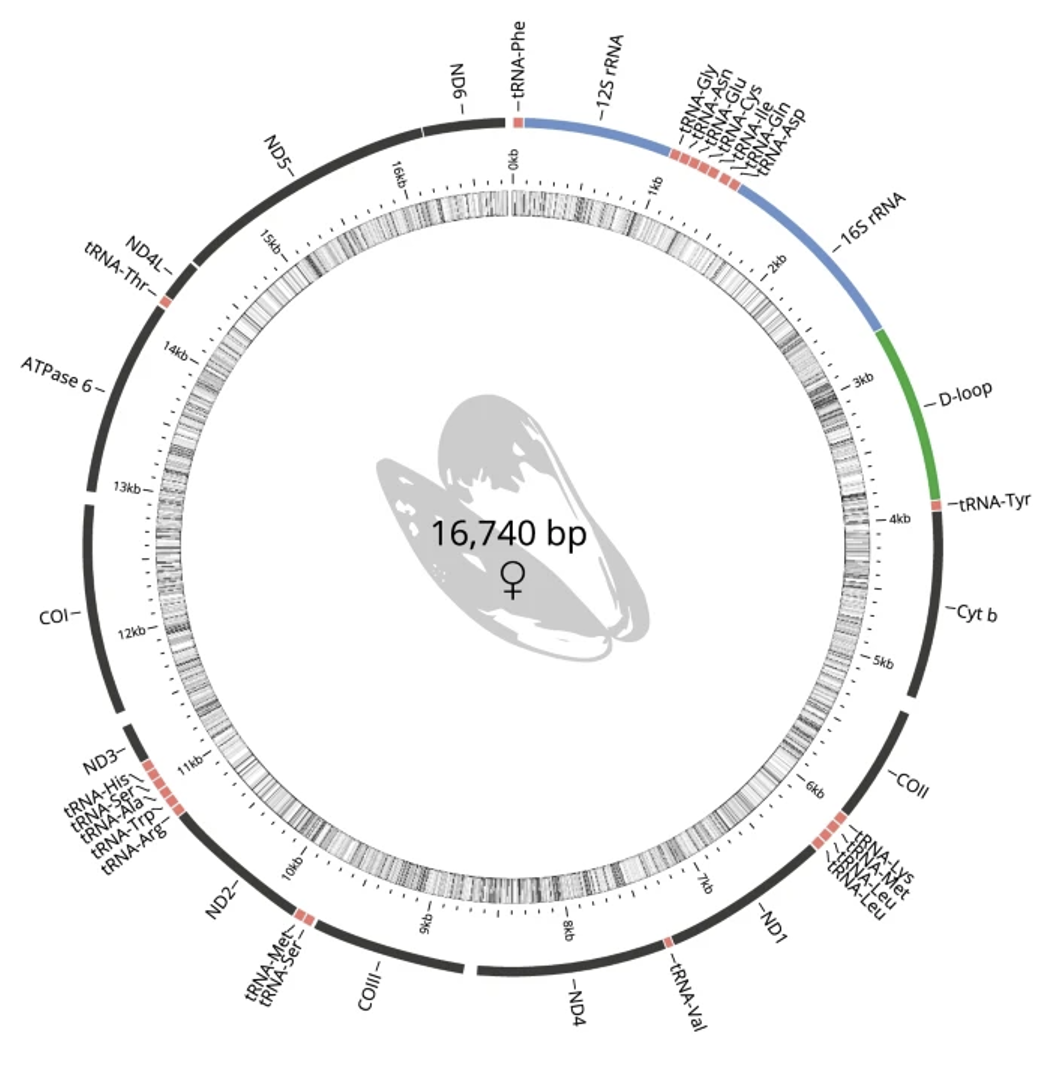
Here are the questions/ chapters we were kicking around last meeting:
The overarching theme is: Exploring xenobiotic effects and consequences in mussels (to mussel physiology)
Chapters 1 & 2 (my MS work) remain the biomarker and DNA methylation work, respectively
Can the mussels from the WDFW Nearshore Monitoring Program be used to assess the impact of contaminants on the mussels using traditional biomarkers?
Can epigenetics (DNA Methylation) be used to identify novel biomarkers of PAH exposure in mussels?
Chapter 3 is lab testing the results of the Chapter 2
Utilize ‘new’ mussels to test the outcomes of the mussels that show differential methylation from Chapter 2.
Short- term (1-3 month) and longer term (3-6 month) lab- based exposure and response experiments designed to characterize the metabolic threshold of mussels exposed to known contaminant mixtures that mimic those in Puget Sound.
- Clarify stress induced DMLs and their pathways. Do they align with the DMLs in Chapter 2?
Chapter 4 is a larger synthesis of the knowledge gained in Chapters 2 and 3.
What are the contaminants inducing in mussels that can have broader population or ecosystem implications?
This can be assessed through a variety of field- based experiments
Outplanting primed and non-primed mussels to assess response at sites with higher or lower contaminant burdens.
Reciprocal outplanting - mussels outplanted at high contaminant sites are moved to sites with low contamination and vice versa.
Larger questions we’re kicking around to use as guides in creating the initial proposal (in addition to those above)
Do epigenetics play a role in creating a phenotypic buffer that supports mussel acclimation/ resilience in the face of climate change drivers (xenobiotic exposure)?
Can we define a tipping- point in mussel’s physiology and/ or their role in ecosystem health?
If there is a tipping point, to what degree does prior exposure create phenotypic response?
What are the ecosystem- level consequences of foundational organisms reaching tipping points?
What mechanisms control phenotypic response to prior exposures (aka epigenetic memory)?
Do we actually have methylation described in mussels? What resources/ information is out there to support putting that picture together?
New (to me) Literature
Reviewing Mytilus trossulus genome literature, I came across a ton of cool papers that I want to discuss in the context of pollutant work, resilience, and hitting a knowledge gap that isn’t a chasm or a dead end. The 3 below are just a few that are interesting to me.
- [Paper 1: Breton et al. (2006) Comparative analysis of gender-associated complete mtDNA genomes in marine mussels] (https://github.com/ChrisMantegna/labnotebook/blob/main/resources/Breton%20et%20al.%20-%202006%20-%20Comparative%20Analysis%20of%20Gender-Associated%20Complete%20Mitochondrial%20Genomes%20in%20Marine%20Mussels%20(%20Myti.pdf)
- Paper 2: Breton et al. (2021) Did doubly uniparental inheritance (DUI) of mtDNA originate as a cytoplasmic male sterility (CMS) system?
- Paper 3: Chelyadina et al. (2022) Effects of heavy metals on sex inversion of the mussel Mytilus galloprovincialis in coastal zone of the Black Sea
Some questions that are coming up
- Do we know why the paternal mtDNA is more degraded than the maternal mtDNA and found dominantly in male offspring?
- Have we seen anything to indicate the population ratios around PS during or after the marine heat waves?
- Is it ethical (assuming they live long enough) to expose mussels to contaminants (heavy metals maybe) induce spawning and sex change and see if there is a difference in methylation patterns?